|
|
-
Takács-Kollár Veronika
Investigation of actin polymerization: methodology and case study
-
P31
Investigation of actin polymerization: methodology and case study
Veronika Takács-Kollár1, Tamás Huber1,2, Péter Gaszler1,2, Réka Pintér1, Rauan Sakenov1, Andrea Teréz Vig1, Mónika Ágnes Tóth1, Beáta Bugyi1,2
1 University of Pécs, Medical School, Department of Biophysics
2 Regional Committee of The Hungarian Academy of Sciences at Pécs, The Expert Committee of Physics and Astronomy, Spectroscopy Committee, Pécs, Hungary
Actin is one of the most important components of the cytoskeleton that plays a crucial role in many cellular processes. The organization of the actin cytoskeleton is under the control of actin binding proteins (ABPs). Total internal reflection microscopy (TIRFM) is a proper technique for the observation and characterization of actin dynamics from single molecules to a complex system in vitro, it works as a bridge between in vitro and in vivo actin polymerization assays. Furthermore, it is suitable to investigate the role of ABPs in modifying the structural and dynamic properties of actin. Flightless-I (Fli-I) is a member of gelsolin superfamily. It has a unique structure including gelsolin-like domains (GH) and leucine-rich repeats (LRR). Fli-I can interact with actin, however, its biochemical activities in actin dynamics regulation are largely elusive.
The aim of our study is to provide an overview of actin polymerization experiments by fluorescence spectroscopy and TIRFM from basic principles to data/image analysis. We test the applicability of different fluorophores and specify the advantages and limitations of each approach. As a case study, we investigate the effect of wild-type Fli-I and its disease-causing mutations and truncations on actin dynamics. Pyrene-actin-based fluorescence spectroscopy reveals a biphasic effect of Fli-I on actin assembly kinetics, complementary TIRFM assays enlightens the molecular mechanism underlying the activities of Fli-I and its disease-related variants at the single filament level.
Acknowledgment
The research in Hungary was funded by NKFIH within the framework of the project TKP2021-EGA-17 and 2021-4.1.2-NEMZ_KI-2022-00025 ” Activities of Flightless I revealed by acto-myosin based in vitro motility techniques” (TH). We thanks to József Mihály (SZBK, Institute of Genetics, Biological Research Centre) for the Fli-I plasmids.
|
|
|
-
Török György
Adaptive changes in the a-band region of the giant protein titin in diseased human cardiac sarcomere
-
P32
Adaptive changes in the A-band region of the giant protein titin in diseased human cardiac sarcomere
György Török1, Iliza Ramazanova1, Péter Dániel1, András Jámbor1, Dalma Kellermayer1,2, Cristina M. Șulea1,2, Zoltán Szabolcs2, Miklós S. Z. Kellermayer1 and Balázs Kiss1
1Department of Biophysics and Radiation Biology, Semmelweis University, Budapest, Hungary
2Heart and Vascular Center, Semmelweis University, Budapest, Hungary
Titin, the largest protein known, spans the half-sarcomere, the contractile unit of skeletal and cardiac muscle through its Z-disk to M-line and interacts with thin and thick filaments in the I- and A-band of the muscle sarcomere, respectively. Titin’s A-band segment is not well understood but is shown to be orders of magnitude less extensible than the I-band region of the molecule. Heterozygous truncating mutations (TTNtv) affecting A-band titin are often associated with dilated cardiomyopathy (DCM). Marfan syndrome (MFS), a connective tissue disorder caused by mutations of the matrix glycoprotein fibrillin is also associated with impaired cardiac contractility but its exact pathomechanism is largely unknown.
Here, we performed STED super-resolution microscopy on sections of stretched and fixed demembranated human cardiac myofibrils carrying heterozygous TTNtv mutations or on samples originated from MFS patients. Sequence-specific anti-titin antibodies included the 1) MIR, which labels titin at the ends of the thick filaments, 2) A170, labeling titin close to the M-line (this epitope is missing in TTNtv+). Sarcomere length-dependent anti-titin epitope position, shape and intensity analysis pointed at structural defects in the I/A junction and the M-band of TTNtv+ sarcomeres. Our experiments indicate that truncated titin is able to integrate into the cardiac sarcomere. We propose that the truncated titin cannot precisely register the ends of the thick filaments, and this can ultimately lead to the manifestation of DCM by disrupting the overlapping of thin and thick filaments. In MFS sarcomeres a pronounced, ~30 nm shift away from the M-line was found in the case of the A170 titin epitope suggesting that alterations in the M-band ultrastructure might be important contributors of the impaired cardiac contractility of MFS patients.
|
|
|
-
Ujfalusi Zoltán
The effects of contrast agents on renal cell lines and on the actin cytoskeleton
-
P33
The effects of contrast agents on renal cell lines and on the actin cytoskeleton
Szilvia Barkó1, Elek Telek1, Kinga Ujfalusi-Pozsonyi1 Gábor Hild1,2 and Zoltán Ujfalusi1
1 Department of Biophysics, Medical School, University of Pécs; Pécs, Hungary
2 Department of Medical Imaging, Clinical Centre, University of Pécs, Pécs, Hungary
Medical images may show some degree of contrast loss in most imaging techniques. In such cases contrast materials are the best tools to enhance the density and intensity of the given area. Nowadays, radiologists can choose from a wide range of contrast agents. The active material of these contrast media penetrates in cells and because of their limited ability of depletion these molecules can accumulate in cells of different tissues. In many cases, contrast agents are used in relatively high volumes, which places a heavy burden on kidney function. A few years ago, contrast agent treatment was identified as the third leading cause of hospital-acquired acute kidney injury (after surgery and hypotension), accounting for 12% of all cases. Today, ~5% of hospitalized patients who develop acute renal failure have normal renal function before contrast administration. We believe that contrast agents exert a significant proportion of their cell-damaging effects by affecting the actin cytoskeleton. The overview of the corresponding literature clarifies that the effects of the clinically applied contrast materials expressed directly on the actin cytoskeleton and its detailed molecular mechanisms are unknown. Our aim is to investigate all possible effects contrast materials can express on human renal cells and the actin protein inside, especially the dynamic organization/rearrangement of the actin network.
Our results clearly show that the applied contrast materials greatly affect the polymerization properties of actin. The examined contrast compounds changed other parameters of actin too and caused dramatic changes on the examined cell lines as well. The DSC results show significantly decreased thermal stability for the treated actin filaments.
|
|
|
-
Vozáry Eszter
Effect of Weight Loss on the Electrical Impedance Parameters of Lettuce and Iceberg Lettuce Stored at Room Temperature
-
P34
Effect of Weight Loss on the Electrical Impedance Parameters of Lettuce and Iceberg Lettuce Stored at Room Temperature
Eszter Vozáry, Bíborka Gillay
Hungarian University of Agriculture and Life Science, Institute of Food Science and Technology
Department of Food Industrial Measurements and Control
We bought the lettuce and iceberg lettuce at the local market. The leaves were stored at room temperature. The weight loss of the leaves was measured with a scale. The electrical impedance spectra were determined with HP4284A and 4285A precision LCR meters in the frequency range of 30 Hz to 30 MHz. At a measuring voltage of 1 V, the magnitude and phase angle of the electrical impedance were measured. The measured spectra were corrected with the stray inductance and capacitance values. A homemade needle electrode and ECG electrodes were used for the measurement. The initial moisture content of the leaves was determined from samples dried to constant weight in an oven at 110°C. Both the impedance measurement and the weight measurement were performed at room temperature for each leaf for 10 minutes over 4-5 hours until the leaf completely withered. The measured impedance spectra were approximated with three distributed elements connected in series. We determined the resistance and capacity of the elements of the model circuit, as well as the relaxation time. The change in the obtained parameters followed well the weight loss, i.e. the change in the water content of the leaf.
|
|
|
-
Balogh Anna
Label-free tracking of cell adhesion kinetics as a function of various parameters
-
P35
Label-free tracking of cell adhesion kinetics as a function of various parameters
Anna Balogh1,2, Kinga Dora Kovacs1,2, Imola Rajmon1,2, Inna Szekacs2,
Beatrix Peter2, Robert Horvath2
1Eötvös Lóránd University, Department of Biological Physics, Budapest, Hungary
2 Nanobiosensorics Laboratory, Institute of Technical Physics and Materials Science, Centre for Energy Research
Most tissue cells cannot survive for more than a few hours without adherence, if we were able to prevent the adhesion of malignant cells, metastasis could be prevented [1] [2]. The data used for analysis was recorded by a surface sensitive, label-free, resonant waveguide grating based optical biosensor. Due to the force calibration of the optical signal provided by the sensor for individual cells, this technique is suitable for determining the force curves of a large number of cells [3]. To analyse the adhesion signals at the molecular level, we applied and developed different kinetic models. These models take into account relevant molecular parameters, such as dissociation and association rates, the two dimensional kinetic dissociation constant which describes the integrin-ligand binding strength [4].
We investigated the extent to which the adhesion kinetics of cells are affected by the surface density of the cells, the density of echistatin, and the effect of gold nanoparticles on cell adhesion. Echistatin is a potent inhibitor of αIIβ3, αvβ3 and α5β1 receptors [5]. Functionalized nanoparticles can penetrate into living cells, Peter et. al. demonstrated from the recorded kinetic adhesion data that the uptake of the functionalized nanoparticles is an active process [6]. Peter et. al. also investigated the role of glycocalyx components in the cellular uptake of nanoparticles [7]. Subjecting these results to further analysis, we developed procedures and codes that enable the study of molecular parameters influencing adhesion in living cells, in their real environment, without isolating the relevant molecules. Our results potentially open the way for further analysis of the kinetic data obtained from the adhering cells.
Acknowledgment
This work was supported by the National Research, Development, and Innovation Office (Grant Numbers: PD 134195 for Z.Sz, PD 131543 for B.P., ELKH topic-fund, "Élvonal" KKP_19 TKP2022-EGA-04 grants).
References
[1] Sudhakar A. History of Cancer, Ancient and Modern Treatment Methods. J Cancer Sci Ther. 1, 2 (2009). https://doi.org/10.4172/1948-5956.100000e2
[2] Lodish, H., Berk, A., Matsudaira, P., et al. Molecular Cell Biology. (2003)
[3] Sztilkovics, M., Gerecsei, T., Peter, B. et al. Single-cell adhesion force kinetics of cell populations from combined label-free optical biosensor and robotic fluidic force microscopy. Sci Rep 10, 61 (2020). https://doi.org/10.1038/s41598-019-56898-7
[4] Kanyo, N., Kovacs, K.D., Saftics, A. et al. Glycocalyx regulates the strength and kinetics of cancer cell adhesion revealed by biophysical models based on high resolution label-free optical data. Sci Rep 10, 22422 (2020). https://doi.org/10.1038/s41598- 020-80033-6
[5] Szekacs, I., Orgovan, N., Peter, B., et. al. Receptor specific adhesion assay for the quantification of integrin–ligand interactions in intact cells using a microplate based, label-free optical biosensor. Sensors and Actuators B: Chemical, 256 (2018). https://doi.org/10.1016/j.snb.2017.09.208
[6] Peter, B., Lagzi, I., Teraji, S., et. al. Interaction of Positively Charged Gold Nanoparticles with Cancer Cells Monitored by an in Situ Label-Free Optical Biosensor and Transmission Electron Microscopy. ACS Appl. Mater. Interfaces 10, 32 (2018). https://doi.org/10.1021/acsami.8b01546
[7] Peter, B., Kanyo, N., Kovacs, K.D., et. al Glycocalyx Components Detune the Cellular Uptake of Gold Nanoparticles in a Size- and Charge-Dependent Manner. ACS Appl. Bio Mater. 6, 1 (2022). https://doi.org/10.1021/acsabm.2c00595
|
|
|
-
Farkasné Bebesi Tímea
Spectroscopic study of extracellular vesicles using plasmonic gold nanoparticles
-
P36
Spectroscopic study of extracellular vesicles using plasmonic gold nanoparticles
Tímea Bebesi Farkasné1,2, Marcell Pálmai1, Imola Csilla Szigyártó1, Anikó Gaál1, Orsolya Bálint-Hakkel3, Attila Bóta1, Zoltán Varga1, Judith Mihály1
1 Research Centre for Natural Sciences, Institute if Material and Environmental Sciences
2 Eötvös Lóránd University, Hevesy György PhD School of Chemistry
3 Centre for Energy Research, Institute of Technical Physics and Material Sciences
Extracellular vesicles (EVs), spontaneously released by cells, play an important role in intercellular communication. Due to their special size and composition (lipid bilayer-bounded nanosystems, usually smaller than 200 nm, containing both proteins and RNA), they play diagnostic, prognostic and therapeutic roles, for example, they can be "new generation" biomarkers of various diseases.
IR spectroscopy, especially attenuated total reflection (ATR), is rapidly emerging as a label-free promising tool for molecular profiling of EVs. However, the relative low number of extracellular vesicles (~1010 particle/mL) and possible impurities (protein aggregates, lipoproteins, buffer molecules, etc.) present in EV samples might result in poor signal-to-noise (S/N) ratio. The plasmonic properties of gold nanoparticles (AuNPs) are used in many characterization techniques, inclusive characterization and testing of EVs. Surface-enhanced infrared spectroscopy (SEIRA – Surface-enhanced IR absorption) using plasmonic nanoparticle, however, is still an unexploited method.
Nanosized gold nanoparticles and tailored nanostructures with confined electromagnetic near-fields were prepared, characterized and tested with model-EVs (EV-like liposomes) and red blood cell derived EVs. A concentration dependent interaction was established between the citrate-stabilized gold nanoparticles and the lipid bilayers, which strongly affected both the plasmonic behaviour of AuNPs and the bilayers lipid organization. At appropriate extracellular vesicle – gold nanoparticle ratio a 6-fold maximum enhancement was obtained in the lipid spectral signatures. Exploiting the fine details of EV – gold nanoparticles interaction, further surface modifications of gold nanoobjects are planned, enhancing the sensitivity and specificity of EV detection enabling a strong platform for IR spectroscopic investigations of EVs.
Acknowledgment
This work was funded by ÚNKP-22-3-II-ELTE-507 and NKFIH K-131657, K131594, 2020-1-1-2- PIACI-KFI_2020-00021, TKP2021-EGA and KKP_22-144180 grants. ZV and MP are supported by the János Bolyai Research Scholarship of the HAS.
|
|
|
-
Biró-Gyuris Katinka
Role of the hHv1 proton channel in vascular smooth muscle cells
-
P37
Role of the hHv1 proton channel in vascular smooth muscle cells
Katinka Gyuris1, Geraldo Domingos1, Éva Korpos2 and Zoltán Varga1
1 University of Debrecen, Faculty of Medicine, Department of Biophysics and Cell Biology
2 University of Debrecen, Faculty of Medicine, Department of Biophysics and Cell Biology, MTA-DE Cell Biology and Signaling Research Group
The hHv1 voltage-dependent proton channel is a passive transporter that selectively transfers protons across the membrane, and thus plays an important role in pH regulation of many cell types. Vascular smooth muscle cells (VSMCs) found in the arterial wall have a resting or contractile phenotype under physiological conditions. When the vessel wall is damaged, the cells switch to a synthetic, migratory and proliferative phenotype, which allows tissue regeneration. Failure of migrating/proliferating cells to switch back to a contractile phenotype induces pathogenic vascular remodelling leading to atherosclerosis.
In our work, we aim to demonstrate the presence of hHv1 on vascular smooth muscle cells and to elucidate the role of the channel in normal and pathological cellular activities through the regulation of intracellular pH. We investigate the role of hHv1 in VSMC survival, differentiation and matrix production.
If hHv1 plays a key role in the pathological activity of VSMCs during atherosclerosis but not in normal function, the channel may become an important pharmacological target to inhibit the pathological activity of VSMCs in atherosclerosis.
To confirm this hypothesis, we performed viability assays using MTT assay, motility experiments using scratch assay method and impedance measurement, PCR and Western blot experiments to detect hHv1, alizarin red staining to visualize the effect on calcified extracellular matrix production, and microscopic studies.
Our results support the hypothesis that hHv1 is expressed in VSMCs and plays a role in their ability to migrate and influences their viability. This conclusion may provide a basis for further experiments to quantify the difference in hHv1 expression between contractile and differentiated proliferative cells and to identify related functional differences.
|
|
|
-
Bóta Attila
Nanoerythrosome-based promising drug delivery system
-
P38
Nanoerythrosome-based promising drug delivery systems
Attila Bóta1, Judith Mihály1, Kinga Ilyés1, Bence Fehér2, Tünde Juhász3, András Wacha1, Heinz Amenitsch4 and Zoltán Varga1
1Research Centre for Natural Sciences, Biological Nanochemistry Research Group, Budapest
2Laboratory of Self-Organizing Soft Matter, Department of Chemical Engineering and Chemistry, Eindhoven University of Technology, Eindhoven, The Netherlands
3Research Centre for Natural Sciences, Biomolecular Self-assembly Research Group, Budapest
4Austrian SAXS beamline@ELETTRA, Are Science Park, Basovizza TS, Trieste, Italy and Inorganic Chemistry, Graz University of Technology, Graz, Austria
Nanoerythrosomes are artificial vesicle-like objects formed from erythrocyte-membranes, named ghosts, by physical processes, such as extrusion or sonication. Phosphatidylcholines (PCs) and sphingomyelins (SMs) are outer membrane constituents, while phosphatidylserines (PSs) and phosphatidylethanolamine (PEs) generally take place on the inner side of the membrane-bilayer. By addition of different artificial lipids, very different size-ranges of nanoerythrosomes can be achieved, therefore proper reference materials and drug delivery systems with adequate surface chemical behaviour can be prepared [1]. The presence of dipalmitoyl-phosphatidylethanolamine (DPPE) results in the formation of larger nanoerythrosomes, while the addition of dipalmitoylphosphatidylcholine (DPPC) induces the formation in a middle-range (140 -160 nm). The presence of the mixture of DPPC - LPC (lysophosphatidylcholine) causes bicelle – micelle type nanoparticles [2]. Here we show that in the complex physic-chemical study, among the different experimental methods (transmission electron-microscopy combined with freeze-fracture (FF-TEM), Microfluidic Resistive Pulse Sensing (MRPS), dynamic light scattering (DLS), the small-angle X-ray scattering (SAXS) turned out to be a powerful tool in the complex physic-chemical study of this drug delivery system.
Acknowledgment
The project was supported by the National Research, Development and Innovation Office of Hungary under grants K131657 (A. Bóta) and K131594 (J. Mihály) and 2018-1.2.1-NKP-2018-00005 under the 2018-1.2.1-NKP funding scheme (A. Bóta, Z. Varga). Z Varga and A. Wacha are supported by the János Bolyai Research Scholarship of the HAS.
References
[1] Deák R, Mihály J, Szigyártó ICS, Beke-Somfai T, Turiák L, Drahos L, Wacha A, Bóta A and Varga Z (2020) Mat. Sci. and Eng. C 109:110428-110437.
[2] Bóta A, Fehér B, Wacha A, Juhász T, Szabó D, Turiák L, Gaál A, Varga Z, Amenitsch H and Mihály J(2023) J Mol. Liq. 369: 120791-120800.
|
|
|
-
Czigléczki Janka Zsófia
Functionally important C-terminus of small GTPase Ran: exploring its nucleotide-specific conformational surface
-
P39
Functionally important C-terminus of small GTPase Ran: exploring its nucleotide-specific conformational surface
Janka Czigleczki1, Pedro Tulio de Resende Lara2, Balint Dudas1,3,4, Hyunbum Jang5, David Perahia4, Ruth Nussinov 5,6and Erika Balog1
1Department of Biophysics and Radiation Biology, Semmelweis University, Budapest, Hungary
2Department of Medical Genetics and Genomic Medicine, School of Medical Sciences, University of Campinas—UNICAMP, Campinas, Brazil
3Inserm U1268 MCTR, CiTCoM UMR 8038 CNRS—Université Paris Cité, Paris, France
4Laboratoire et Biologie et Pharmacologie Appliquée, Ecole Normale Supérieure Paris-Saclay, Gif-sur-Yvette, France
5Computational Structural Biology Section, Frederick National Laboratory for Cancer Research in the Cancer Innovation Laboratory, National Cancer Institute, Frederick, MD, United States
6Department of Human Molecular Genetics and Biochemistry, Sackler School of Medicine, Tel Aviv University, Tel Aviv, Israel
As a member of the Ras superfamily of small GTPases, Ran (Ras-related Nuclear protein) is the main regulator of the nucleo-cytoplasmic transport through the nuclear core complex. It functions as a molecular switch cycling between the GDP-bound inactive or “off” and GTP-bound active or “on” state. Since deregulation of Ran is linked to numerous cancers from the stage of cancer initiation to metastasis, understanding the complexity of its interaction, especially the regulatory mechanism, is critical for drug discovery.
Ran consists of a globular (G) domain and a C-terminal region, which is bound to the G-domain in the inactive, GDP-bound states. The crystal structures of the GTP-bound active form complexed with Ran binding proteins (RanBP) show that the C-terminus undergoes a large conformational change, embracing Ran binding domains (RanBD), whereas in the crystal structures of macromolecular complexes not containing RanBDs the structure of the C-terminal segment remains unresolved, indicating its large conformational flexibility. This movement could not have been followed either by experimental or simulation methods. Here, by using molecular dynamics (MD) and MDeNM (Molecular Dynamics with excited Normal Modes) simulation methods, we present how rigid the C-terminal region is in the inactive RanGDP form and for the first time in the literature, we were able to follow its conformational flexibility in the GTP-bound form. This conformational mapping allows us to envisage how the C-terminus can embrace RanBDs during the function of Ran.
The simulations were carried out by JC, and were analyzed and interpreted by JC, PR, BD, HJ, RN, DP, and EB.
|
|
|
-
Domingos Geraldo Jorge
Identification of inhibitors of the human hv1 proton channel
-
P40
Identification of inhibitors of the human hv1 proton channel
Geraldo Domingos1, Adam Feher1, Eva Korpos 1,2, Tibor G. Szanto1, Martina Piga3, Tihomir Tomasic3, Nace Zidar3, Adrienn Gyongyosi4, Judit Kallai4, Arpad Lanyi4, Ferenc Papp1, Katinka Gyuris1, Zoltan Varga1
1 Department of Biophysics and Cell Biology, Faculty of Medicine, University of Debrecen, Hungary,
2 MTA-DE Cell Biology and Signalling Research Group, Faculty of Medicine, University of Debrecen, Hungary
3 Department of Pharmaceutical Chemistry, University of Ljubljana, Slovenia, 4 Department of Immunology, Faculty of Medicine, University of Debrecen, Hungary
The human voltage-gated proton channel (hHV1) plays an important role in immune system and cancer cells being involved in functions such as proliferation, migration, and oxidative burst. HV1 does not have a conventional ion-conducting pore, the conduction occurs through the voltage-sensing domain. This difference may be the reason for the lack of selective hHV1 inhibitors. Currently, 5-chloro-2-guanidinobenzimidazole (ClGBI) is the most widely used inhibitor of HV1 but it has low selectivity for the channel. This could lead to misinterpretation of functional assays addressing the role of HV1 with the use of ClGBI. Thus, our aim was to find potent and more selective inhibitors for hHV1, which could be useful research tools and serve as lead molecules for the development of drug molecules targeting HV1.
We used patch-clamp to test the affinity and selectivity of potential inhibitors of HV1 on CHO and HEK cells expressing hHV1 and other channels. Seven “hit” molecules were identified among the NZ family of compounds of which NZ-13 has the best selectivity profile.
The widely-used HV1 inhibitor ClGBI blocks various ion channels and therefore is not a selective HV1 blocker. This must be considered in functional tests investigating the role of HV1 in healthy and pathological conditions.
We have identified a new family of hHV1 inhibitors, which have comparable affinities for the channel to ClGBI.
Most NZ molecules have low selectivity for hHV1, but NZ-13, the one with the highest selectivity, may be better suited for functional tests than ClGBI as it inhibits T cell proliferation less.
|
|
|
-
Farkas Enikő
Controlling Live Cell Adhesion through Characterization of Biofunctionalized Surfaces using Label-Free Biosensors
-
P41
Controlling Live Cell Adhesion through Characterization of Biofunctionalized Surfaces using Label-Free Biosensors
Eniko Farkas1, Kinga Dóra Kovács1,3, Beatrix Peter1, Attila Bonyár2, Sandor Kurunczi1, Inna Szekacs1, and Robert Horvath1
1 Nanobiosensorics Laboratory, Institute of Technical Physics and Materials Science, Centre for Energy Research, Budapest, Hungary
2 Department of Electronics Technology, Faculty of Electrical Engineering and Informatics, Budapest University of Technology and Economics, Budapest, Hungary
Biomaterial coatings that possess cell-repellent or cell-adhesive properties have a significant interest in medical and biotechnological applications [1-4]. However, conventional approaches lack in-depth analysis and quantitative comparison of these coatings for regulating adhesion, particularly for bacterial cell adhesion. Label-free Optical Waveguide Lightmode Spectroscopy (OWLS) can offer a solution for the detailed analysis of biomaterial coatings. OWLS biosensors detect the optical properties of the adhesive surface using evanescent waves with a penetration depth of 100-150 nm [5-7]. This surface-sensitive technique enables a thorough evaluation of biomaterial coatings for regulating adhesion. Uniquely, OWLS enables the in situ measurement of both the coating process and subsequent cell adhesion.
The present study utilizes the OWLS method for in-depth characterization of biomaterial surfaces with regard to bacterial adhesion. Initially, adhesion blocking biomaterials, namely bovine serum albumin, I-block, PAcrAM-g-(PMOXA, NH2, Si), (PAcrAM-P), and PLL-g-PEG, with varying coating temperatures, were screened. PAcrAM-P exhibited the best blocking capability against bacterial concentrations up to 107 cells/mL. Subsequently, different immobilization methods, such as Mix&Go (AnteoBind) films, protein A, avidin-biotin based surface chemistries, and simple physisorption, were employed to captureEscherichia coli specific antibodies. Bacterial cell adhesion was then tested on immobilized antibodies with various blocking agents. The OWLS analysis allowed for the determination of the parameters of the applied agents by considering the kinetic data of adhesion, the surface mass density, and the protein orientation. Based on the experimental results, surfaces were created and tested for controlling both bacterial and mammalian cell adhesion. [8]
Acknowledgment
This work was supported by the "Lendület" (HAS) research program, the National Research, Development and Innovation Office of Hungary ((ERC_HU, VEKOP 2.2.1-16, ELKH topic-fund, "Élvonal" KKP_19 and KH grants, PD 131543 and TKP2022-EGA-04 –INBIOM TKP Programs financed from the NRDI Fund). This work was also supported by 77 Elektronika Ltd. by their supplying of antibodies and reagents.
References
[1] Frutiger A, et. al. (2021) Chem Rev 121: 8095–8160.
[2] Rigo S, et. al. (2018) Adv Sci 5: 1700892.
[3] Castillo-Henríquez L, et. al. (2020) Sensors 20: 6926.
[4] D’Agata R, et. al. (2021) Polymers 13:1929.
[5] Vörös J, et. al. (2002) Biomaterials 23: 3699–3710.
[6] Tiefenthaler K, et. al. (1989) J Opt Soc Am. B 6: 209–220.
[7] Saftics A, et. al. (2021) Adv Colloid Interface Sci 294: 102431–102433.
[8] Farkas E, et. al. (2022) Biosensors 12: 56.
|
|
|
-
Ghofrane Medyouni
Role of ion channels in CAR T-cell
-
P42
Role of ion channels in CAR T-cell
Ghofrane Medyouni1, Vivien Jusztus1, Orsolya Vörös1, Maria Eduarda Lima1,2, György Panyi1, Péter B. Hajdu1, 2
1 University of Debrecen, Faculty of Medicine, Department of Biophysics and Cell Biology
2 University of Debrecen, Faculty of Dentistry, Division of Dental Biochemistry
Cancer immunotherapy partly relies on the reprogramming of host immune cells to eliminate cancer cells. Genetic modification of T cells to express chimeric antigen receptors (CARs) is utilized in the treatment of hematological malignancies. Despite its success, many challenges remain to improve the efficacy and safety of this therapy. Ion channels in T-cells participate in the regulation of Ca2+-dependent activation pathway and play a role in various effector functions inevitable for target cell abolition. Hence, modification of ion channels’ function can contribute to successful immune therapy. However, no study has been reported about functional role of CAR T-cell ion channels yet.
We established a 3rd-generation CAR expressing cell line (CD19-CAR cells) from Jurkat cells. We used the whole-cell patch-clamp technique and FURA-2-based Ca2+-imaging to determine the biophysical properties of Kv1.3 and Ca2+-response of CD19-CAR cells, respectively. We adapted a Calcein Red based killing assay to test CD19-CAR cells’ target cell killing capacity. We assessed the localization of Kv1.3 in standalone and in the CAR-synapse engaged CD19-CAR cells.
We showed that Kv1.3 activation and inactivation kinetics are the same in non-transfected and CD19-CAR cells, while voltage-dependence of activation was different. Thapsigargin-induced Ca2+-response of CD19-CAR cells was lower as compared to the control. We showed that Kv1.3 channels are co-localized with CARs in standalone CD19-CAR cells, and they redistribute to the contact region between a CD19-CAR cell and a target cell (Raji B cell). Upon Vm24 addition (specific Kv1.3 inhibitor, 1 nM) the target cell ability of CD19-CAR cells was impaired. Based on these results, we suppose that ion channels can affect the outcome of immunotherapy, and further experiments are needed to clarify their functional role.
Acknowledgment
This work was supported by Stipendium Hungaricum Scholarship to M.G.t and NKFIH (K128525, P.H.).
|
|
|
-
Jusztus Vivien
Ion channel expression of CD8+ T cells in ovarian cancer
-
P43
Ion channel expression of CD8+ T cells in ovarian cancer
Vivien Jusztus1, Ghofrane Medyouni1, Orsolya Vörös1, Zsolt Szabó1, Rudolf Lampé3, György Panyi1, Orsolya Matolay3, Eszter Maka3, Zoárd Krasznai3, Péter Hajdu1,2
1 University of Debrecen, Faculty of Medicine, Department of Biophysics and Cell Biology
2 University of Debrecen, Faculty of Dentistry, Division of Dental Biochemistry
3 University of Debrecen, Faculty of Medicine, Department of Gynecology and Obstetrics
The ion channels of T lymphocytes have an important role in effector functions such as activation, cytokine production and tumor cell elimination. T cells recognise and kill cancer cells during continuous monitoring. Although tumor infiltrating lymphocytes (TILs) are able to penetrate the tumor, they cannot fulfil their effector function due to the suppressive nature of the tumour microenvironment. K+ channels, such as Kv1.3 and KCa3.1, stabilize the negative membrane potential of T cells to control Ca2+-influx through CRAC channels and Ca2+-dependent signaling. In the present study, we determined the expression of T cell ion channels from peripheral blood of untreated ovarian cancer patients and healthy donors.
PBMCs were isolated from blood of ovarian patients and healthy donors using Ficoll-Paque density gradient method. Cells were activated with CD3/CD28 antibodies. Whole-cell current was measured in activated CD8+ T cells using patch-clamp technique. Ca2+ response of CD8+ cells was evaluated with FURA-2 Ca2+-imaging method.
KCa3.1 expression level in blood CD8+ cells from malignant tumor patients were lower than in healthy and benign tumor groups. Contrary, the Kv1.3 conductance of CD8+ T cells were significantly higher in malignant tumor patients as compared to other two groups. To asses Ca2+ response of cells, we determined the quotient of FURA-2 ratios measured in 2 mM Ca2+ and 0 mM Ca2+ after thapsigargin addition: there was no differences between the groups.
In summary, we suppose that down-regulation of KCa3.1 expression and Kv1.3 level upregulation in blood CD8+ cells could be a reporter on the presence of malignancy, as we reported before for head and neck cancer patients [1].
Acknowledgment
This work was supported by Stipendium Hungaricum Scholarship to M.G. and NKFIH (K128525, P.H.).
References
[1] Chimote AA, Balajthy A, Arnold MJ, Newton HS, Hajdu P, Qualtieri J, et al. (2018) Sci Signal. 11(527): 1–12
|
|
|
-
Kashmala Shakeel
Isolation of novel peptide toxins from the venom of the scorpion Centruroides bonito which block Kv1.2 ion channel with picomolar affinity
-
P44
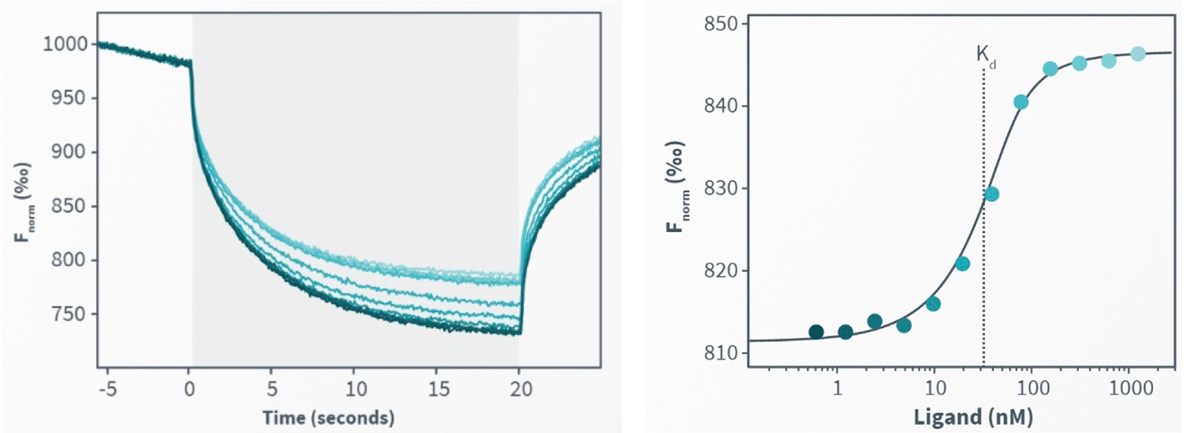
Isolation of novel peptide toxins from the venom of the scorpion Centruroides bonito which blocks Kv1.2 ion channel with picomolar affinity
Kashmala Shakeel1, Muhammad Umair Naseem1, Lourival D Possani2, Gyorgy Panyi1
1 Department of Biophysics and Cell Biology, Faculty of Medicine, University of Debrecen, Hungary
2 Departamento de Medicina Molecular y Bioprocesos, Universidad Nacional Autónoma de México, Mexico
Seven new peptide toxins named as CboK1 to CboK7 were isolated from the venom of the Mexican scorpion Centruroides bonito by liquid chromatography. The primary structure of these peptides were determined by Edman degradation. Mass spectrometry analysis was used to determine the molecular weights which range between 3760.4 Da to 4357.9 Da, comprising 32 to 39 amino acid residues cross-linked with three tightly folded disulfide-bridges. The amino acid sequence alignment with known potassium scorpion toxins (KTx) and phylogenetic tree analysis unveiled that CboK1 (α-KTx 10.5) and CboK2 (α-KTx 10.6) belong to α-KTx 10 subfamily, whereas CboK3 (α-KTx 2.22), CboK4 (α-KTx 2.23), CboK6 (α-KTx 2.21), CboK7 (α-KTx 2.24) bears more than 95% amino acid similarity with the members of α-KTx 2 subfamily, and CboK5 is 100% identical with previously described Ce3 toxin (α-KTx 2.10). The electrophysiological assays (whole-cell patch clamp) revealed that except CboK1, all other six peptide toxins blocked the voltage-gated potassium channel Kv1.2 with high affinity, having Kd values in the picomolar range (24-763 pM) and inhibited the Kv1.3 ion channel with comparatively less potency (Kd values between 20-171 nM). Moreover, CboK2 and CboK3 inhibited ~10% and CboK7 inhibited ~50% of Kv1.1 currents at 100 nM concentration. Among all CboK7 (α-KTx 2.24) has the highest affinity for Kv1.2 ion channel with Kd value of 24 pM, and reasonable selectivity over Kv1.3 (~1000-fold) and Kv1.1 (~6000-fold) ion channels. These distinguishable characteristics of the CboK7 toxin may provide a framework for developing tools to treat Kv1.2-related gain of function channelopathies.
|
|
|
-
Lambrev Petar
Light energy harvesting by photosystem I in cyanobacterial cells
-
P45
Light energy harvesting by photosystem I in cyanobacterial cells
Petar Lambrev1, Parveen Akhtar1, Avratanu Biswas1, Ivo van Stokkum2
1 Biological Research Centre, Szeged, Institute of Plant Biology
2 Universiteit Amsterdam, 1081 HV Amsterdam, The Netherlands
Photosystem I (PSI) is a crucial component of the light-dependent reactions of oxygenic photosynthesis occurring in cyanobacteria, algae and plants. It is a multi-subunit pigment-protein complex binding more than a hundred pigment – chlorophylls (Chls) and carotenoids, as well as cofactors carrying out photoinduced electron transport with a quantum yield of near unity. Whereas PSI in plants and eukaryotic algae is attached to peripheral membrane-intrinsic light-harvesting complexes, cyanobacteria utilize the membrane extrinsic phycobilisomes (PBS) as the main light-harvesting antenna complex1. While the structural and energetic interaction between the PBS and photosystem II (PSII) is well established, less is known about the connectivity of PBS and PSI and the ability of the PBS to transfer energy directly to PSI is debated. We investigated the pathways and dynamics of energy transfer from PBS to the photosystems in Synechocystis sp. PCC 6803. The excitation kinetics of PBS and PSI were followed by picosecond time-resolved fluorescence spectroscopy in the wild-type strain and in a mutant devoid of PSII2. We found that PBS are capable of directly transferring energy to PSI in the PSII-deficient mutant, in a time scale of about 20 ps at room temperature. Based on an earlier model of energy transfer in Synechocystis sp. PCC 68033 and simultaneous fitting to the measured data of isolated complexes and intact cells, a detailed model of energy transfer between different PBS, PSI and PSII chromophore groups was obtained.
Many cyanobacterial species, when exposed to iron limitation conditions, produce a specialized pigment-protein complex, IsiA, that is known to associate into rings around PSI4. The physiological function of IsiA is not fully understood. In isolated PSI-IsiA complexes IsiA efficiently transfers absorbed photon energy to the PSI core5, which can extend the absorption cross-section of the photosystem and help reduce the number of iron-rich PSI core complexes in the cells. However, IsiA has also been proposed to have a photoprotective, energy-dissipating role or to serve as a Chl depot6. To find more about the light-harvesting role of IsiA in vivo, we followed the cellular content of IsiA in cells of Synechocystis sp. PCC 6803 under iron limitation and investigated the energy transfer from IsiA to PSI by time-resolved spectroscopy. IsiA formed PSI-IsiA supercomplexes in vivo having similar energy transfer characteristics as isolated supercomplexes – confirming the primary role of IsiA as an accessory light-harvesting antenna to PSI. However, a significant fraction (40%) remained unconnected to PSI, supporting the notion of a dual functional role of IsiA. Moreover, we found that Synechocystis mutants containing only monomeric PSI contained far fewer IsiA units per PSI compared to the wild-type strain. We conclude that the trimeric organization of PSI in wild-type Synechocystishas role both in the accumulation and the energy transfer capabilities of IsiA under iron stress.
References
1. Blankenship, R. E., Molecular mechanisms of photosynthesis. John Wiley & Sons: 2021.
2. Bittersmann, E.; Vermaas, W., Fluorescence lifetime studies of cyanobacterial photosystem II mutants. Biochim. Biophys. Acta 1991, 1098 (1), 105–116.
3. van Stokkum, I. H.; Gwizdala, M.; Tian, L.; Snellenburg, J. J.; van Grondelle, R.; van Amerongen, H.; Berera, R., A functional compartmental model of the Synechocystis PCC 6803 phycobilisome. Photosynth. Res. 2018, 135 (1-3), 87–102.
4. Toporik, H.; Li, J.; Williams, D.; Chiu, P.-L.; Mazor, Y., The structure of the stress-induced photosystem I–IsiA antenna supercomplex. Nat. Struct. Mol. Biol. 2019, 26 (6), 443–449.
5. Andrizhiyevskaya, E. G.; Frolov, D.; van Grondelle, R.; Dekker, J. P., Energy transfer and trapping in the photosystem I complex of Synechococcus PCC 7942 and in its supercomplex with IsiA. Biochim. Biophys. Acta 2004, 1656 (2–3), 104–113.
6. Jia, A.; Zheng, Y.; Chen, H.; Wang, Q., Regulation and functional complexity of the chlorophyll-binding protein IsiA. Frontiers in Microbiology 2021, 12, 774107.
|
|
|
-
Magyaródi Beatrix
Role of glycocalyx in cancer cell adhesion: kinetics of interactions from label-free optical biosensor measurements
-
P46
Role of glycocalyx in cancer cell adhesion: kinetics of interactions from label-free optical biosensor measurements
Beatrix Magyaródi1, Boglárka Kovács1, Inna Székács1, Robert Horvath1
1 Nanobiosensorics Laboratory, Research Centre for Energy Research, Institute for Technical Physics and Materials Science, Konkoly-Thege u 29-33, 1120 Budapest, Hungary
The glycocalyx is a sugar rich layer covering the surface of the cells.[1] It is composed of glycoproteins and proteoglycans. The cellular glycocalyx plays an important, but not yet understood, role in cellular signaling and metabolism, its disorders generate pathological process.[2] Interestingly, the thickness of the glycocalyx layer of cancer cells is significantly larger compared to that of healthy cells. This fact further highlights the importance of glycocalyx in tumor progression and treatment. In an earlier work, a regulatory mechanism of cellular glycocalyx in cancer adhesion was revealed using label-free optical biosensor, fluorescent microscopy, and cell surface charge measurements. [3]
The primary goal of our work is to study the role of glycocalyx components in cellular adhesion by employing various types of digesting methods. In these initial measurements we use a label-free, high-throughput, resonant waveguide grating-based optical biosensor. The instrument is well suited for monitoring of cellular adhesion kinetics in real-time, even at the single-cell level.[4]
Acknowledgment
This work was supported by the National Research, Development, and Innovation Office (Grant Numbers: PD 134195 for Z.Sz, PD 131543 for B.P., ELKH topic-fund, "Élvonal" KKP_19 TKP2022-EGA-04 grants)
References
[1] M. J. Paszek et al., “The cancer glycocalyx mechanically primes integrin-mediated growth and survival,” Nature, vol. 511, no. 7509, pp. 319–325, 2014, doi: 10.1038/nature13535.
[2] E. R. Cruz-Chu, A. Malafeev, T. Pajarskas, I. V. Pivkin, and P. Koumoutsakos, “Structure and response to flow of the glycocalyx layer,” Biophys. J., 2014, doi: 10.1016/j.bpj.2013.09.060.
[3] N. Kanyo et al., “Glycocalyx regulates the strength and kinetics of cancer cell adhesion revealed by biophysical models based on high resolution label-free optical data,” Sci. Rep., 2020, doi: 10.1038/s41598-020-80033-6.
[4] M. Sztilkovics et al., “Single-cell adhesion force kinetics of cell populations from combined label-free optical biosensor and robotic fluidic force microscopy,” Sci. Rep., 2020, doi: 10.1038/s41598-019-56898-7.
|
|
|
-
Solymosi Katalin
The effect of light and age of the leaves on plastid differentiation and essential oil composition of spearmint (Mentha spicata L.)
-
P47
The effect of light and age of the leaves on plastid differentiation and essential oil composition of spearmint (Mentha spicata L.)
Adrienn Dobi1, Anna Skribanek2, Bernadett Szögi-Tatár3, Andrea Böszörményi3, Imre Boldizsár1, and Katalin Solymosi1
1Eötvös Loránd University, Budapest, Hungary
2Eötvös Loránd University, Savaria University Centre, Szombathely, Hungary
3Institute of Pharmacognosy, Semmelweis University, Budapest, Hungary
Spearmint (Mentha spicata L.) is a widely used spice, aromatic and medicinal plant with a characteristic fragrance. Its valuable active substances are monoterpene and sesquiterpene essential oil components produced by exogenous secretory structures, i.e. the various glandular hairs located on the shoot, especially on the leaves. The biosynthesis of the terpenoid essential oils requires isoprenoid biosynthesis of peculiar plastids present in the glandular hairs.
In this work, we have investigated 1) whether the prolamellar body-like membrane structures observed in the neck cells of peltate glandular hairs of spearmint can be considered as homologues with the prolamellar bodies present in etioplasts of dark-grown plants, 2) whether the presence and the activity (essential oil production) of the glandular hairs depends on the developmental stage of the leaves, 3) whether light conditions (e.g. light or dark development) affect plastid differentiation and the essential oil production of the leaves.
We have described in detail the various plastids and the organization of their inner membranes in the different cells of the exogenous secretory structures of spearmint, i.e. in capitate and peltate glandular hairs. We found clear structural differences between the prolamellar bodies of spearmint etioplasts of dark-grown leaves and of the similar structures found in secretory cells, which rather ressembled regularly arranged clusters of plastoglobuli, and not tubuloreticular membranes. Dark-growth did not influence much the structure of the plastids in the secretory cells, and our data obtained with solid-phase microextraction followed by gas chromatography and mass spectrometry (GC/MS) confirmed that not the illumination conditions but rather the developmental stage of the leaves influences the essential oil composition of spearmint.
Acknowledgments
The work was supported by the ÚNKP-22-5 New National Excellence Program of the Ministry for Culture and Innovation from the source of the National Research, Development and Innovation Fund (to K.S.) and by the Bolyai János Research Scholarship of the Hungarian Academy of Sciences (to K.S.).
|
|
|
-
Muhammad Umair Naseem
Cm39 (α-KTx 4.8): A novel scorpion toxin that inhibits voltage-gated K+ channel Kv1.2 and small- and intermediate-conductance Ca2+-activated K+ channels KCa2.2 and KCa3.1
-
P48
Cm39 (α-KTx 4.8): A novel scorpion toxin that inhibits voltage-gated K+ channel Kv1.2 and small- and intermediate-conductance Ca2+-activated K+ channels KCa2.2 and KCa3.1
Muhammad Umair Naseem1, Georgina Gurrola-Briones2, Margarita R. Romero-Imbachi3, Jesus Borrego1, Edson Carcamo-Noriega2, José Beltrán-Vidal3, Fernando Z. Zamudio2, Kashmala Shakeel1, Lourival D. Possani2, Gyorgy Panyi1
1 University of Debrecen, Faculty of Medicine, Department of Biophysics and Cell Biology, Hungary
2 Departamento de Medicina Molecular y Bioprocesos, Universidad Nacional Autónoma de México, Mexico
3 Departamento de Biología, Facultad de Ciencias Naturales, Universidad del Cauca, Colombia
A novel peptide toxin, Cm39, was identified in the venom of the Colombian scorpion Centruroides margaritatus. It is composed of 37 amino acid residues with a MW of 3980.2 Da and folded by three disulfide bonds. The Cm39 sequence also contains the Lys-Tyr (KY) functional dyad required to block voltage-gated K+ (Kv) channel. Amino acid sequence comparison with previously known K+ channel inhibitor scorpion toxins (KTx) and phylogenetic analysis revealed that Cm39 is a new member of α-KTx 4 family and registered with systematic number of α-KTx4.8. The full chemical synthesis and proper folding of Cm39 was obtained. The pharmacological properties of the synthetic peptide were determined using patch-clamp electrophysiology. Cm39 inhibits the voltage-gated K+ channel hKv1.2 with high affinity (Kd = 65 nM). The conductance-voltage relationship of Kv1.2 was not altered in the presence of Cm39, the analysis of the toxin binding kinetics was consistent with a bimolecular interaction between the peptide and the channel, and therefore the pore blocking mechanism is proposed for the toxin-channel interaction. Cm39 also inhibits the Ca2+-activated KCa2.2 and KCa3.1 channels, with Kd = 575 nM, and Kd = 59 nM, respectively, however, the peptide does not inhibit hKv1.1, hKv1.3, hKv1.4, hKv1.5, hKv1.6, hKv11.1, mKCa1.1 potassium channels or the hNav1.5 and hNav1.4 sodium channels at 1 µM concentration. Understanding the unusual selectivity profile of Cm39 motivates further experiments to reveal novel interactions with the vestibule of toxin-sensitive channels [1].
References
[1] Naseem MU, Gurrola-Briones G, Romero-Imbachi MR, Borrego J, Carcamo-Noriega E, Beltrán-Vidal J, Zamudio FZ, Shakeel K, Possani LD and Panyi G, (2023) Toxins 15(1), p.41.
|
|
|
-
Papp Ferenc
A synthetic flavonoid derivate modulates the fluorescent signal of voltage-gated proton channels
-
P49
A synthetic flavonoid derivate modulates the fluorescent signal of voltage-gated proton channels
Zoltán Pethő1,2, Gilman E. S. Toombes, Dávid Pajtás1, Martina Piga3, Zsuzsanna Magyar4, Nace Zidar3, György Panyi1, Zoltán Varga1 and Ferenc Papp1
1 Department of Biophysics and Cell Biology,Faculty of Medicine, University of Debrecen,
2 Institut für Physiologie II, Münster, Germany
3 Department of Pharmaceutical Chemistry, Faculty of Pharmacy, University of Ljubljana
4 Department of Physiology, Faculty of Medicine, University of Debrecen
The voltage-sensing domain (VSD) of voltage-gated proton channel (Hv1) serves as a pore for protons as well as a voltage sensor, which makes this channel unique among voltage-gated channels. Natural flavonoids, which are widely distributed and act as chemical messengers and physiological regulators in plants, modulate the function of some voltage-gated ion channels (EAG1, HCN2. etc.) in animal cells. We have designed synthetic flavonoid derivatives to inhibit the current of Hv1. We produced and tested tens of flavonoid derivatives on Ciona intestinalis Hv1 using the voltage-clamp fluorometry (VCF). The most potent compound, molecule #109, changed the originally negative VCF signal to positive and altered the biphasic VCF signal shape to monophasic. Also, this molecule caused a rightward shift in the conductance-voltage relationship in a concentration dependent manner. This flavonoid derivative quenched the TAMRA-MTS fluorescence in cuvette and on frog oocytes decreasing the baseline fluorescence, independently the oocyte expressed Hv1 or not. Furthermore, #109 could stain the membrane of HEK cells. These results indicate that #109 binds not directly to CiHv1 but to the cell membrane and in this way, it indirectly modifies the gating of Hv1 and the VCF signal, as a strong quenching molecule in close vicinity of the fluorophore. The latter was confirmed by our model calculations, in which we assumed that molecule #109, as a strong quencher, is embedded in the cell membrane close to TAMRA and during the conformational change due to depolarization, TAMRA is continuously moving away from this strong quencher molecule. Therefore, the VCF signal becomes a continuously increasing fluorescence change from the originally complex shape, which was overall a decreasing fluorescence change.
Acknowledgment
This work was supported by OTKA Bridging Fund 1G3DBKB0BFPF247 (FP); by János Bolyai Research Scholarship of the Hungarian Academy of Sciences (BO/00355/21/8) (FP); by the ÚNKP-21-5-DE-460 New National Excellence Program of the Ministry for Innovation and Technology (FP); OTKA 132906 (ZV). This study was also funded by the Slovenian Research Agency (Grant No. P1-0208) (NZ).
|
|
|
-
Patty, C.H. Lucas
Remote detection of life through full-Stokes spectropolarimetry
-
P50
Remote detection of life through full-Stokes spectropolarimetry
C.H. Lucas Patty1, Mathilda Fatton2, Urs A. Schroffenegger1, Saskia Bindschedler2, Pilar Junier2, Brice-Olivier Demory1
1Center for Space and Habitability (CSH), Universität Bern, Switzerland
2Institute of Biology, University of Neuchâtel, Switzerland
Homochirality is a generic and unique property of all biochemical life as we know it. There is a growing concensus that homochirality is a universal prerequisite of life and therefore as a biosignature free from the assumptions made by using terrestrial life as a benchmark [1]. Spectropolarimetry and, in particular, the detection of non-zero signatures in circular polarization is an indicator of the homochiral nature of molecular and supramolecular organic matter and is thus a direct and intuitive proxy for the remote detection of life using unpolarized incident light such as from the Sun [2]. We will describe the ongoing effort to characterize and quantify the nature of these chiroptical signals resulting from living organisms. While various studies have been performed to this end on eukaryotic photosynthetic organisms, in both the laboratory and in the field, including aerial observations (see e.g. [3][4]), relatively little systematic observations have been made of prokaryotic life. The results gathered so far on microbial mats and pure cultures show a remarkable variety in terms of both polarimetric spectral shape and magnitude [5]. Within the framework of the SenseLife project, we aim to further characterize and quantify the nature of these signals including the polarimetric signal response to external factors and physiology. We will further present novel results demonstrating the potential of characterization aerobiology using spectropolarimetry in the visible. Within this context we will describe the performance of FlyPol [2], a fast and sensitive spectropolarimetric instrument dedicated to the remote detection of linear and circular polarizance. In addition, we will present the results of ongoing aerial and field measurement campaigns probing the polarizance resulting from natural habitats, providing an outlook on the endeavor of measuring especially circular polarizance from space using Earth as a benchmark.
1. Glavin, D. P., Burton, A. S., Elsila, J. E., Aponte, J. C., & Dworkin, J. P. (2019). The search for chiral asymmetry as a potential biosignature in our solar system. Chemical reviews, 120(11), 4660-4689.
2. Patty, C. H. L., Ten Kate, I. L., Sparks, W. B., & Snik, F. (2018). Remote sensing of homochirality: a proxy for the detection of extraterrestrial life. In Chiral Analysis (pp. 29-69). Elsevier.
3. Patty, C. H. L., Pommerol, A., Kühn, J. G., Demory, B. O., & Thomas, N. (2022). Directional aspects of vegetation linear and circular polarization biosignatures. Astrobiology, 22(9), 1034-1046.
4. Patty, C. H. L., Kühn, J. G., Lambrev, P. H., Spadaccia, S., Hoeijmakers, H. J., Keller, C., ... & Demory, B. O. (2021). Biosignatures of the Earth-I. Airborne spectropolarimetric detection of photosynthetic life. Astronomy & Astrophysics, 651, A68.
5. Sparks, W. B., Parenteau, M. N., Blankenship, R. E., Germer, T. A., Patty, C. H. L., Bott, K. M., ... & Meadows, V. S. (2021).
|
|
|
-
Polgár Tamás Ferenc
Assessing the effect of photobleaching on morphometric parameters of different cell types in the central nervous system
-
P51
Assessing the effect of photobleaching on morphometric parameters of different cell types in the central nervous system
Tamás F. Polgár1,2, Krisztina Spisák1,2, Zalán Kádár1, Nora Alodah3, László Siklós1 and Roland Patai1
1 Institute of Biophysics, Biological Research Centre, Szeged, Hungary
2 Theoretical Medicine Doctorate School, University of Szeged, Szeged, Hungary
3 College of Medicine, Alfaisal University, Kingdom of Saud Arabia
The sensitivity of immunofluorescence to the illumination time and intensity is well known; however, its effects on morphological parameters are less considered. Morphometric evaluation of different cell types could indicate the condition of the region of interest; for example, thickness and number of branches in microglia can imply their activation state.
Fluorescent detection using immunohistochemistry (IHC) targeting microglia/macrophages (fine structures) and neurons (bulk structures) was performed on neural tissues using different types of fluorophores while IHC with photostable diamonibenzidine was served as standard. Time series (0.5, 1, 2, 5, 10 and 15 minutes of illumination after taking the first image) were acquired from each spinal cord sections with the initial microscope settings. After standard mean intensity measurements, morphological parameters were extracted. Dynamic and relative total area microglial density were measured with a macro developed in our lab used routinely for quantifying glial activation. Fractal geometrical parameters were measured with the help of the Fiji plugin FracLac, and changes were examined between the initial image and the images containing faded structures.
Standard mean intensity measurements show different fading properties of different fluorophores. Dynamic and relative total area density comparisons show that in some cases all structures can fade to the point of becoming non-detectable after 2 minutes of illumination, while in some fractal geometrical parameters, more than 35 % differences can be observed.
Our results suggest that while fluorescent detection using IHC is an excellent method for localization and co-localization, for proper fine structure morphological measurements a photostable staining method is essential.
Acknowledgment
This work was partially supported by the Ministry for National Economy of Hungary through the GINOP-2.3.2-15-2016-00034 program. K.S. was supported from the UNKP-21-3-SZTE-73 New National Excellence Program of the Hungarian Ministry of Human Capacities. T.F.P. was supported by the EFOP 3.6.3-VEKOP-16-2017-00009 with the financial aid of the Ministry of Finance. R.P. was supported by the “National Talent Programme” with the financial aid of the Ministry of Human Capacities (NTP-NFTÖ-21-B-0203).
|
|
|
-
Sallai Igor
Label-free immune cell analysis using optical biosensor
-
P52
Label-free immune cell analysis using optical biosensor
I. Sallai1, Z. Szittner1, Sz. Novák1, I. Székács1 and R. Horvath1
1 Nanobiosensorics Laboratory, Center of Energy Research, Eötvös Loránd Research Network, Budapest, Hungary
Understanding of activation processes at the single-cell level in response to different stimuli is essential for the diagnosis of certain diseases.
External stimuli induced differences are reflected in the dynamic changes of cell biophysical parameters, such as cell motility, shape, spreading and adhesion properties [1]. Novel highly sensitive optical biosensors allow the monitoring of changes in these parameters in a label-free manner. The advantage of label-free detection is that cells can be examined in an intact/organism-specific manner without modification.
The above parameters can be studied using microplate-based, high-throughput systems [2]. Surface functionalisation provide the opportunity to create an organism-specific environment [3]. In classical measurements, such as microscopy, dye-conjugated antibody labelling is essential and can be combined with label-free data [4].
Our aim is to interpret the activation mechanisms of various cell types and their function triggered by different stimuli and compare the results with conventional testing methods.
Acknowledgment
This work was supported by the National Research, Development, and Innovation Office (Grant Numbers: PD 134195 for Z.Sz, ELKH topic-fund, "Élvonal" KKP_19 TKP2022-EGA-04 grants).
References
[1] Z. Szittner, B. Péter, S. Kurunczi, I. Székács, and R. Horvath, “Functional blood cell analysis by label-free biosensors and single-cell technologies,” Advances in Colloid and Interface Science, vol. 308. Elsevier B.V., Oct. 01, 2022. doi: 10.1016/j.cis.2022.102727.
[2] M. Sztilkovics et al., “Single-cell adhesion force kinetics of cell populations from combined label free optical biosensor and robotic fluidic force microscopy,” Sci Rep, vol. 10, no. 1, Dec. 2020, doi: 10.1038/s41598-019-56898-7.
[3] N. Orgovan et al., “In-situ and label-free optical monitoring of the adhesion and spreading of primary monocytes isolated from human blood: Dependence on serum concentration levels,” Biosens Bioelectron, vol. 54, pp. 339–344, Apr. 2014, doi: 10.1016/j.bios.2013.10.076.
[4] S. Zheng, J. C. H. Lin, H. L. Kasdan, and Y. C. Tai, “Fluorescent labeling, sensing, and differentiation of leukocytes from undiluted whole blood samples,” Sens Actuators B Chem, vol. 132, no. 2, pp. 558–567, Jun. 2008, doi: 10.1016/j.snb.2007.11.031.
|
|
|
-
Sóti Adél
Effect of salt stress on etioplast and chloroplast membranes of thylakoid transporter mutants of Arabidopsis thaliana
-
P53
Effect of salt stress on etioplast and chloroplast membranes of thylakoid transporter mutants of Arabidopsis thaliana
Helga Fanni Schubert 1, Adél Sóti 1, Richard Hembrom 1, Roumaissa Ounoki 1, Enkhjin Enkhbileg1, Emilija Dukic 2, Cornelia Spetea 2, and Katalin Solymosi 1
1 Eötvös Loránd University, Budapest
2 University of Gothenburg, Gothenburg, Sweden
Soil salinity is an increasing problem for agriculture worldwide. Salinity has a complex effect on plants and influences the structure of plastids in different ways. Most often the effect of salt stress is studied in leaf chloroplasts, and in several cases swelling of the intrathylakoidal space of chloroplast inner membranes is reported under such conditions. However, it is yet unclear what causes the swelling of these membranes, and whether it has any relation to ion transport processes across these membranes. In this work, plastid ultrastructure was compared in the cotyledons and leaves of Arabidopsis thaliana plants of different developmental stages and grown under different light regimes under control conditions as well as under salt stress (30 min treatment with 200 or 300 mM NaCl or 600 mM NaCl:KCl, 1:1). In addition to the wild-type (WT) plants, we also analysed the thylakoid membrane structure and photosynthetic activity in single, double and triple mutants of the thylakoid-located voltage-gated chloride ion channel VCCN1, chloride ion channel CLCe and potassium proton exchanger KEA3. The above salt treatment did not affect the structure of the photosynthetic apparatus of mature chloroplasts in old leaves, however, it influenced the structure of chloroplasts in cotyledons in various ways, indicating the sensitivity of the young seedlings to the stress, and the potential presence of protective mechanisms that stabilize chloroplast structure at later developmental stages even under the above stress conditions. Salt stress also had an effect on the etioplasts of both WT and some mutant plants.
Acknowledgment
The work was funded by the grant OTKA FK124748, and supported by the ÚNKP-22-5 New National Excellence Program of the Ministry for Culture and Innovation from the source of the National Research, Development and Innovation Fund (to K.S.) and by the Bolyai János Research Scholarship of the Hungarian Academy of Sciences (to K.S.).
|
|
|
-
Szatmári Tímea
Effect of rational modification of disordered domains of the epidermal growth factor receptor on its biophysical characteristics
-
P54
Effect of rational modification of disordered domains of the epidermal growth factor receptor on its biophysical characteristics
Tímea Szatmári1, Péter Nagy1
1 Department of Biophysics and Cell Biology, Faculty of Medicine, University of Debrecen
The epidermal growth factor receptor (EGFR) belongs to the ErbB receptor tyrosine kinase family. The extracellular domain of EGFR consists of four subdomains (I-IV). Upon ligand binding, the extracellular (EC) domain of the receptor is rearranged, resulting in exposition of the dimerization arm of subunit II providing an opportunity to form homodimers or heterodimers. The 3D structure determined by the amino acid sequence affects protein function. Proteins in which the formation of an ordered structure is not complete are called intrinsically disordered proteins. Recently, an algorithm (FuzPred) was developed that predicts the tendency of proteins to form disordered regions and their structural changes upon interacting with other proteins. The amino acid sequence of the EGFR was analyzed by FuzPred, which determines the tendency of distinct sequence regions to constitute globular (assuming ordered conformations), disordered (intrinsically disordered adopting ordered conformation upon binding), or fuzzy regions (remaining disordered even in their bound state). The following mutations were designed based on the software prediction that alter the fuzziness and the molecular interactions of EGFR domain II. 1. Mutation T274G is predicted to result in increased dynamics and destabilization of the dimerization site. It still forms a dimer, but the interaction is weak. 2. Mutation Q276F is expected to decrease dynamics of the dimerization arm and create a rigid binding site. As a result, it is unable to form a dimer due to lack of flexibility. 3. Mutation K284Q is expected to decrease dynamics while strengthening the binding site. This mutation promotes dimerization. We successfully generated all three mutants (dark and EGFP tagged on the C terminal of the proteins) by site-directed mutagenesis and stably transfected CHO cells expressing the EGFP-tagged mutant EGFRs. We investigated the ligand binding and the cooperativity of the wild-type EGFR and its mutant variants. CHO cells transfected with the wild-type or the mutant EGFRs were labeled with a concentration series of fluorescently labeled EGF (TAMRA-EGF). Our results suggest that those mutants that were predicted to be less prone to dimerization bind EGF less cooperatively and with a slightly lower affinity. In our further experiment we would like to examine the dimerization process (FRET, N&B) itself and consequent transmembrane signaling.
|
|
|
-
Szikszainé Ritter Zsuzsanna
Analysis of unilateral Walker A and A-loop mutants indicate that a single active catalytic site is sufficient to promote transport in ABCB1
-
P55
Analysis of unilateral Walker A and A-loop mutants indicate that a single active catalytic site is sufficient to promote transport in ABCB1
Zsuzsanna Ritter1,2, Szabolcs Tarapcsák1, Zsuzsanna Gyöngy1,2, Orsolya Bársony1, Nimrah Ghaffar1,2 Thomas Stockner3, Gergely Szakács4, and Katalin Goda1
1Department of Biophysics and Cell Biology, Faculty of Medicine, University of Debrecen, Debrecen, Hungary
2Doctoral School of Molecular Cell and Immune Biology, University of Debrecen, Debrecen, Hungary
3Institute of Pharmacology, Center for Physiology and Pharmacology,
4Institute of Cancer Research, Medical University of Vienna, Vienna, Austria
The human ABCB1 is a full transporter with two nucleotide binding domains (NBDs) and two pseudo-symmetric transmembrane domains (TMDs). The two NBDs form two symmetrically arranged composite nucleotide binding sites (NBSs). Each NBS is formed by the A-loop, H-loop, Walker A, Walker B and Q-loop of one NBD, and the X-loop and signature sequence of the other NBD. The conserved tyrosine of the A-loop aligns the adenine ring of the bound ATP, contributing to nucleotide binding affinity through stacking interactions. The Walker A lysine interacts with the α and β phosphate of ATP. The two nucleotide binding domains were shown to be functionally equivalent, and the integrity of both catalytic centers is generally believed to be needed for transport. Consistently with the widely accepted models predicting that the two NBDs hydrolyze ATP in a strictly alternating order, unilateral mutation of these residues have been described to disrupt ATP hydrolysis and even affect ATP binding.
Here we demonstrate that while ABCB1 variants carrying bilateral A-loop or Walker A mutations are completely inactive, the unilateral exchange of the A-loop tyrosine to alanine or the unilateral mutation of the Walker A lysine to methionine is compatible with both ATP hydrolytic activity and transport function. Characterization of the single mutants revealed the significant (about 10-fold) reduction of the apparent ATP binding affinity compared to wild-type ABCB1. Stabilization of the post-hydrolytic complex by phosphate mimicking anions, such as vanadate or BeFx also occurred at higher ATP concentrations compared to wild-type, supporting that the mutated site probably has an effect on the overall conformation of the NBD dimer. Although the basal catalytic activity was strongly reduced in accordance with the decreased ATP binding affinity of the single mutants, the degree of ATPase stimulation by verapamil was almost identical to that of the wild-type, showing that drug-stimulation of the ATPase activity is preserved in the single mutants. Location of the mutation in the N or C terminal NBD did not affect the extent of ATPase stimulation by verapamil. Taken together, our data indicate that, in contrast to prevailing views, single-site NBD mutant ABCB1
|
|
|
-
Tárnoki-Zách Júlia
Evaluation of peptide carrier candidates using tissue barrier models
-
P56
Evaluation of peptide carrier candidates using tissue barrier models
Júlia Tárnoki-Zách1, Bence Stipsicz2,3, Előd Méhes1, Ildikó Szabó2,4, Kata Horváti4, Bernadett Pályi5, Zoltán Kis5, Szilvia Bősze3,5 András Czirók1
1 Department of Biological Physics, Eötvös Loránd University, Pázmány Péter sétány 1/A, H-1117, Budapest, Hungary
2 ELKH-ELTE Research Group of Peptide Chemistry, Eötvös Loránd Research Network, Eötvös Loránd University, Pázmány Péter sétány 1/A, H-1117, Budapest, Hungary
3 Doctoral School of Biology, Eötvös Loránd University, Pázmány Péter
sétány 1/C, H-1117, Budapest, Hungary
4 MTA-TTK Lendület Peptide-Based Vaccines Research Group, Institute of Materials and Environmental Chemistry, Research Centre for Natural Sciences, Budapest H-1117, Hungary.
5 National Public Health Center, Albert Flórián út 2-6, Budapest, 1097, Hungary
Targeting peptides represent a promising approach to improve uptake and efficiency of pharmacological compounds. In vitro barrier models are valuable screening tools to evaluate peptide transport, uptake and toxicity. Here we characterize a number of readily available lung and kidney epithelial cell lines in a transwell barrier model. After forming a monolayer and a subsequent maturation phase of cell-type specific duration, the epithelial cells develop tight junctions on the surface of polycarbonate inserts as evidenced by beta-catenin and ZO1 immunolabeling as well as delayed transport of targeting peptides. Daily monitoring of transepithelial electrical resistance (TEER) values reveal a cell line specific, characteristic time course. Thus, the TEER method with this calibration data offers a non-destructive and agent-free procedure to time pharmacological transport measurements, and to evaluate cytotoxicity of the transported agents. To characterize peptide targeting efficiency, a detector cell layer was cultured in the basolateral compartment of the barrier tissue model. Uptake of fluorescein-labelled peptides was evaluated by flow cytometry. We demonstrate that a derivative of the well-known peptide penetratin that also contains the neuropilin-binding sequence from tuftsin, a naturally occurring tetrapeptide produced by enzymatic cleavage from immunoglobulin G, is better able to pass through barrier layers that expresses its receptor, NRP-1.
|
|
|
-
Tóth Ádám Viktor
Molecular characterization of pathological and tissue-specific TRPM2 cation channel variants
-
P57
Molecular characterization of pathological and tissue-specific TRPM2 cation channel variants
Ádám V. Tóth1, Ádám Bartók1 and László Csanády1
1 Department of Biochemistry, Semmelweis University, Budapest, Hungary
TRPM2 is a temperature-sensitive, Ca2+-permeable, non-selective cation channel, showing high level of expression in cells of the central nervous system, bone marrow, granulocytes and pancreatic β-cells. Activation of the channel requires the simultaneous intracellular presence of adenosine diphosphate ribose (ADPR), Ca2+ ions and phosphatidylinositol 4,5-bisphosphate (PIP2). In these cells, TRPM2 contributes to Ca2+ influx resulting in important physiological and pathological functions, such as body temperature regulation, cytokine production, oxidative stress response, inflammation or controlled cell death. Moreover, certain TRPM2 point mutations show close genetic connection with bipolar disorder (D543E, R755C) [1] or amyotrophic lateral sclerosis and Parkinson's dementia (P1018L) [2]. Interestingly, alternative splice products were isolated from healthy neutrophil granulocytes (ΔC-TRPM2) [3] and from striatum (SSF-TRPM2) [4], which presumably modify the ligand specificity and function of the channel in a cell-specific manner.
Until now, the mentioned variants have only been investigated using fluorescent imaging techniques or whole-cell electrophysiological methods providing limited opportunities to study the ligands acting intracellularly. Our aim is to examine the listed ion channel variants in molecular details. To this end, expression vector encoding TRPM2 variants have been produced and expressed transiently in HEK cells. Functional measurements are performed by inside-out patch clamp configuration enabling reliable recordings of micro- and macroscopic currents and fast exchange of intracellular ligands. With our method, it is possible to map crucial biological and biophysical parameters of the channel variants: ADPR and Ca2+ sensitivity, gating parameters, inactivation kinetics, temperature dependence. This detailed knowledge is essential for a comprehensive understanding of the role of these mutants in pathomechanisms and tissue-specific variant functions.
References
[1] A. McQuillin et al. Mol Psychiatry 11, 134-142 (2006)
[2] M. C. Hermosura et al. Proc Natl Acad Sci USA 105, 18029-34 (2008)
[3] E. Wehage et al. J Biol Chem 277, 23150-6 (2002)
[4] T. Uemura et al. Biochem Biophys Res Commun 328, 1232-43 (2005)
|
|
|
-
Tóth Dániel
Substrate selectivity of Sulfotransferase Isoenzymes, results based on Molecular Dynamics and Ensemble Docking
-
P58
Substrate selectivity of Sulfotransferase Isoenzymes, results based on Molecular Dynamics and Ensemble Docking
Dániel Toth1,2 , Bálint Dudas2,3, David Perahia3, Erika Balog1, Maria A. Miteva2
1Department of Biophysics and Radiation Biology, Semmelweis University, Hungary
2Inserm U1268 MCTR, CiTCoM UMR 8038 CNRS - Université Paris Cité, France
3Laboratoire de biologie et pharmacologie appliquee, Ecole Normale Superieure Paris-Saclay, France
Sulfotransferase enzymes (SULTs) are a family of cytosolic globular proteins in the chain of metabolism. By catalysing a sulfate transfer from their co-factor, 3′-Phosphoadenosine 5′-Phosphosulfate (PAPS), they eliminate a large variety of small molecules like drugs, hormones and neurotransmitters. Even though the tertiary structure across the family is very similar, their substrates vary considerably in size and complexity. The aim of our project is to better understand the reasons of selectivity between the different SULT isoenzymes, by comparing the broad targeting hepatic detoxifier SULT1A1, and the ileum located, dopamine selective SULT1A3.
Based on our previous results and Molecular Dynamics (MD) and Molecular Dynamics with excited Normal Modes (MDeNM), an extended conformational space of the PAPS-bound SULT1A1 was explored. Further developments of our method utilising ensemble docking with categorised ligands, a method known as Virtual Screening was achieved. Moreover, we have broadened our scope to use the same approach for the SULT1A3.
Based on our new results, we identified the key differences, that are responsible for changing the protein dynamics and binding mechanisms, by opening the binding pocket to an unfavourable conformation for the most common ligands of 1A1, thus acting as efficient selectors. These results can be helpful in the future to develop an algorithm for machine learning, that could differentiate and even predict new substrates of the different isoforms, thus helping in the development of ADME-Tox profiling of novel drug candidates and xenobiotics.
Acknowledgment
TKP2021 EGA 23, Ministry for Innovation and Technology in Hungary,
2021-0143339, Ministére francais de l’Europe et des Affaires Etrangéres
Tét-Balaton project „drug-drug interactions of sulfotransferases” ( 2019-2.1.11-TÉT-2020-00096 )
|
|
|
-
Tóth Gabriella
Examinations of cellular uptake of cell penetrating peptides in vitro and in vivo
-
P59
Examinations of cellular uptake of cell penetrating peptides in vitro and in vivo
Gabriella Tóth1, Gyula Batta1,2, Levente Kárpáti3, Árpád Szöőr1, István Mándity3, Péter Nagy1
1University of Debrecen, Faculty of Medicine, Department of Biophysics and Cell Biology
2University of Debrecen, Faculty of Science and Technology, Institute of Biotechnology, Department of Genetics and Applied Microbiology
3Semmelweis University, Faculty of Pharmacy, Institute of Organic Chemistry
Cell-penetrating peptides (CPPs) are peptides that enter cells by endocytosis and/or directly through the cell membrane. CPPs in general have been considered potential carriers of molecules that have difficulties entering cells. This is the feature that we would like to exploit and thereby establishing the opportunity for CPPs to have therapeutic applications in the long term. Our previously published results have shown that we can increase the cellular uptake and endosomal release of CPPs with statins. Our goal was to modify them and test if it is possible to make them enter the cells more efficiently. We also aimed to test the biodistribution of CPPs in mice after intravenous administration. We examined the cellular uptake and endosomal release by flow cytometry and confocal microscopy in SKBR-3 and MDA-MB-231 cell lines, while for the in vivo experiments a mouse model was applied. Fluorescently-labeled CPPs were used both in the in vivo and in vitro experiments. We compared the differences in the biophysical properties of the original and the modified CPPs, and we found that the cellular uptake of the modified version is more effective. There is a difference between the enhancement in the uptake of CPPs labeled by the pH-sensitive naphthofluorescein or Alexa Fluor 532. In the case of in vivo experiments, we found that peptides enter the mouse organs, including the liver, for which we have shown that CPPs is present in the intracellular space of hepatocytes. CPPs hold promise for increasing the efficiency and specificity of drug delivery to cells.
|